7 Antimicrobial resistance projecticum
7.1 Introduction
The antimicrobial project is done on behalf of RIVM (Rijksinstituut voor Volksgezondheid en Milieu) for the client Gijs Teunis. During the antimicrobial project, our research group worked on further developing and refining the R shiny app for typing and visualizing bacterial plasmids by adding additional epidemiological data, such as information on isolation date, geographical location, the ability to add and compare own fasta/plasmids with the database, and optimizing for easy uploading. We will also integrate patient metadata and visualise them by making an (interactive) map for instance. Additionally, we want to add user-friendly features that allow users to add isolates and compare them with our dataset. There are many options as to visualisation of patient data [1], for instance a sankey diagram to display the type of antiobitica used for a certain infection.
7.2 About antimicrobial resistance
Antimicrobial resistance (AMR) poses a significant threat to global health as bacteria continually adapt to survive antibiotic treatments. These adaptations involve various mechanisms, including the production of enzymes to neutralize antibiotics, the use of efflux pumps to remove them from cells, and the modification of antibiotic targets. Additionally, bacteria can acquire resistance genes from both natural reservoirs and human activities like extensive antibiotic use in medicine, agriculture, and industry. These genes can spread through horizontal gene transfer, transitioning from environmental bacteria to pathogenic strains, exacerbating clinical resistance issues. Given the gravity of AMR’s impact on public health, understanding its mechanisms and origins is paramount for devising effective strategies to curb antibiotic misuse and limit the spread of resistant genes [2].
Furthermore, antibiotic resistance occurs when bacteria change to resist the effects of antibiotics. This can happen through mutations in bacteria’s own genes or by acquiring new genes from other bacteria. In environments like water and soil, bacteria have a diverse range of genes that can help them survive antibiotics. Over time, these resistant genes can spread between bacteria, making infections harder to treat. Scientists study how resistance develops to find better ways to combat it and develop new treatments [3].
7.2.1 R shiny app
Shiny is a tool in R that changes R code into interactive web apps, combining R’s analysis skills with web development. With Shiny, developers can make lively tools that work on web browsers.With Shiny, developers can make user interfaces and write code in R language. Then, Shiny makes this code into HTML, CSS, and JavaScript for smooth web viewing. Shiny apps use R code on the server, helping with lots of data jobs, from simple math to hard stats and machine learning. Shiny gets user info, does stuff with it in R, and shows results right away. This makes Shiny great for exploring data interactively [4]. The previous research group working on this project, made an R shiny app (see figure 8.1) with gene information, mostly. It is our task to add and visualize patient data, such as patient zipcode, sample data, age, gender etc. Forrtunately, there are many options to visualize clinical data [5].
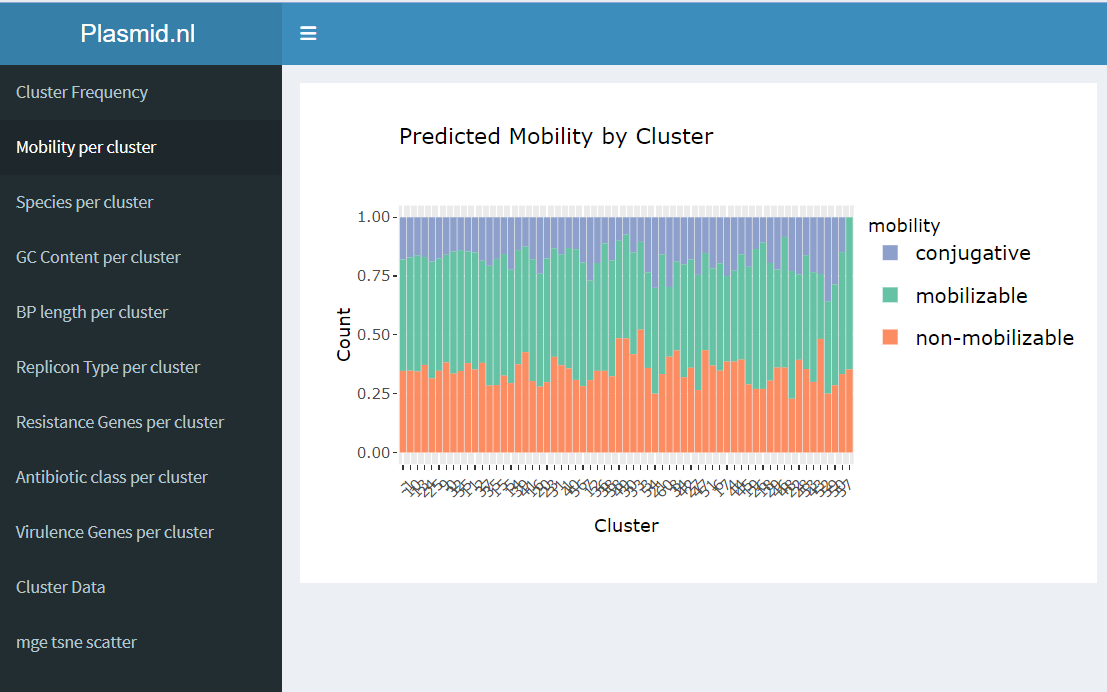
Figure 7.1: The R shiny app.
References
- Appsilon. (2021). Appsilon R Shiny Demo Gallery. https://www.appsilon.com/shiny-demo-gallery
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010 Sep;74(3):417-33. doi: 10.1128/MMBR.00016-10. PMID: 20805405; PMCID: PMC2937522. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2937522/
- Larsson DGJ, Flach CF. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022 May;20(5):257-269. doi: 10.1038/s41579-021-00649-x. Epub 2021 Nov 4. PMID: 34737424; PMCID: PMC8567979. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8567979/
- Domino Data Lab.(2024). Platform.https://www.dominodatalab.com/platform.
- Laure Cougnaud, Michela Pasetto.(2024).Visualization of clinical data. https://cran.r-project.org/web/packages/clinDataReview/vignettes/clinDataReview-dataVisualization.html#visualization-of-summary-statistics